The NCKU OrchidBase
Development and Goals
As one of the largest and most diverse of the angiosperm families [1], the
family Orchidaceae the 25,000–35,000 species are of ecological and evolutionary importance. Like all other living organisms,
present-day orchids have evolved from ancestral forms as a result of selection pressure and adaptation. They
show a wide diversity of epiphytic and terrestrial growth forms and have successfully colonized almost every
habitat on Earth. Specific interaction between orchid flowers and pollinators [2], sequential and rapid interplay
between drift and natural selection [3], the role of obligate orchid–mycorrhizal interactions [4], and epiphytism
may all contribute to the species richness within the Orchidaceae.
The radiation of the orchid family probably took place in a
comparatively short period in comparison to that of most flowering plant
families, which had already started to diversify in the Mid-Cretaceous [5]. The
time of origin of orchids is in dispute, although Dressler suggested that they
originated 80 to 40 million years ago (late Cretaceous to late Eocene). Perhaps
the only general statement that can be made about the origin of orchids is that
most extant groups are probably very young. Recently, the origin of the
Orchidaceae was dated using a fossil orchid and its pollinator; the authors [6]
showed that the most recent common ancestor of extant orchids lived in the late
Cretaceous (76-84 Mya).
According
to molecular phylogenetic studies, Orchidaceae comprise five subfamilies,
including Apostasioideae, Cypripedioideae, Vanilloideae, Orchidoideae and
Epidendroideae.
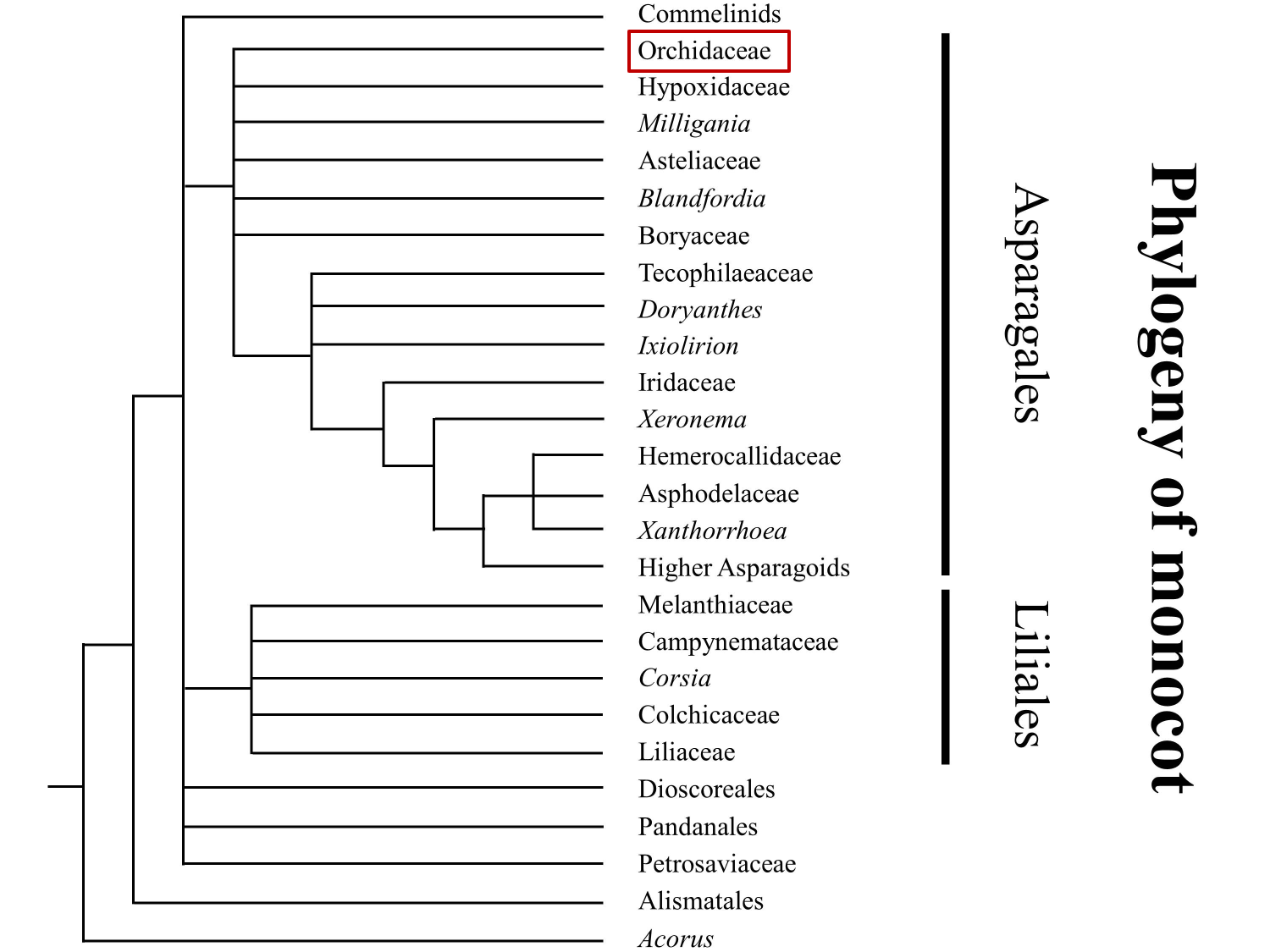

They are known for their diversity of specialized reproductive
and ecological strategies. For successful reproduction, the production of
labellum and gynostemium (a fused structure of androecium and gynoecium) to
facilitate pollination is well documented and the co-evolution of orchid flowers
and pollinators is well known [7-8]. In addition, mature pollen grains packaged
as pollinia, pollination-regulated ovary/ovule development, synchronized timing
of micro- and mega-gametogenesis for effective fertilization, and the release of
thousands or millions of immature embryos (seeds without endosperm) in a mature
capsule may also account for the especially successful evolutionary progress of
orchids [9]. However, despite their unique developmental reproductive biology,
as well as specialized pollination and ecological strategies, orchids remain
under-represented in molecular studies relative to other species-rich plant
families [10].
Orchids are one of the most ecological and evolutionary significant plants, and the Orchidaceae is one
of the most abundant families in angiosperm. The genetic databases will be useful not only for gene discovery
but future genomic annotation. For this purpose, OrchidBase was established for providing sequenced orchid
genome information as well as transcriptomes collected from various tissues of different orchid species.
OrchidBase architecture is composed of a web interface, a SQL Server database management system and
a windows application. The web interface is implemented in static HTML pages and the latest .NET
(Microsoft .NET framework 4.62) software technology. The OrchidBase was developed in part on the basis of
Model-View-Controller (MVC) architecture principles. More specifically, the solution was developed by using
the ASP.NET MVC 4 framework and coded with Visual C# programming language, which is dynamically
executed for querying the database. IIS 6.0 on the Microsoft Windows Server 2016 Standard is adopted for
main system operation. Genome Browser is visualized under Apache web Server on the Ubuntu 16.04. In
addition, based on XML and simple object access protocol (SOAP) Web Services, the system also offers a
web-service interface. For storing and managing collected sequence information and the annotation data, the
SQL Server 2012 system is adopted. The windows application executes a sequence analysis, and Perl and
the C# program are applied to automatically parse data and construct the database. A number of open
source tools and technologies were used for improving database coverage, the user interface and system
performance. The interactive data visualization web page is based on D3 and ASP.NET MVC. For building a
web-based visualization and presenting data in an interactive and convenient way with maximum
compatibility, D3.js, the powerful JavaScript toolkit, was applied to create cross-platform vector graphics. The
JBrowse is applied to navigate orchid genomes. JBrowse is the AJAX-based browsers helping preserve the
user's sense of location by avoiding discontinuous transitions, instead offering smoothly animated panning,
zooming, navigation, and track selection. Consequently, the establishment of OrchidBase will provide
researchers a high-quality genetic resource for data-mining and facilitate efficient experimental studies on
orchid biology and biotechnology.
Reference:
1.Atwood JT:The size
of Orchidaceae and the systematic distribution of epiphytic orchids.Selbyana
1986,9:171-186.
2.Cozzolino S, Widmer
A: Orchid diversity: an evolutionary consequence of deception? Trends in Ecology
&
Evolution 2005, 20:487-494.
3.Tremblay RL,
Ackerman JD, Zimmerman JK, Calvo RN: Variation in sexual reproduction in orchids
and its
evolutionary consequence: a spasmodic journey to diversification.
Biological Journal of the Linnean Society
2005, 84:1-54.
4.Otero JT,
Flanagan NS: Orchid
diversity – beyond deception. Trends in Ecology & Evolution 2006, 21:64-65.
5.Crane PR, Friis EM,
Pedersen KR: The origin and early diversification of angiosperms. Nature 1995,
374:27-33.
6.Ramirez SR,
Gravendeel B, Singer RB, Marshall CR,
Pierce
NE: Dating the origin of the Orchidaceae from
a
fossil orchid with its pollinator.Nature 2007, 448:1042-1045.
7.Yu H, Goh CJ:
Molecular genetics of reproductive biology in orchids. Plant Physiol 2001,
127:1390-1393.
8.Schiestl FP, Peakall
R, Mant JG, Ibarra F, Schulz C, Franke S, Francke W: The chemistry of sexual
deception in an orchid-wasp pollination system.Science 2003, 302:437-438.
9.Tsai WC, Hsiao YY,
Pan ZJ, Kuoh CS, Chen WH, Chen HH: The role of ethylene in orchid ovule
development. Plant Sci 2008, 175:98-105.
10.Peakall R: Speciation in the
Orchidaceae: confronting the challenges. Molecular Ecology 2007, 16:2834-2837.
Comments and suggestions :
Contact Us